In 1965, Margaret Dayhoff published the Atlas of Protein Sequence and Structure, which collated the 65 proteins whose amino acid sequences were then known.
Inspired by that Atlas, today we are releasing the Dayhoff Atlas of protein sequence data and protein language models.
Jul 25, 2025 · 8:14 PM UTC
The Dayhoff Atlas dramatically expands the scale and diversity of publicly available protein data by providing the largest open dataset of natural proteins to date, GigaRef, and a first-in-class, large-scale dataset of synthetic proteins, BackboneRef.
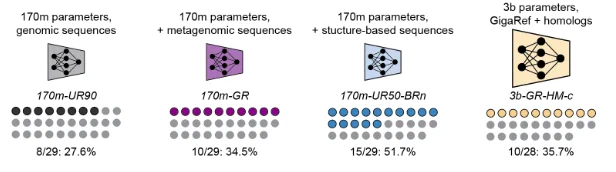
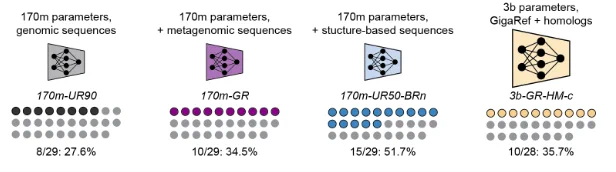
How do dataset choice and model scale affect the quality of proteins generated by the Dayhoff model? In the first study of its kind, we generated sequences from different Dayhoff models and tested them head-to-head in the lab, measuring whether expressed in E. coli.
Learning on GigaRef yielded a small increase in the fraction of expressed proteins. Increasing model and dataset scale further improved the expression rate. Augmenting training with structure-based synthetic data from BackboneRef produced the highest expression success rate.
Our models, code, and data are openly available on Github, Zenodo, and Huggingface.
huggingface.co/collections/m…
zenodo.org/records/15265289
github.com/microsoft/dayhoff
There's a lot more in the preprint!
biorxiv.org/content/10.1101/…
The Dayhoff Atlas was a big team effort, including
@SarahAlamdari
Alex J Lee
Kaeli Kaymak-Loveless
@samir_char
@garykbrixi
@cdomingoenrich
@WChentong
Suyue Lyu
@nfusi
@ntenenz
@avapamini