Bioinformatics Scientist at the Arc Institute. Working on bioinformatics tools for functional genomics and ML in single cell.
San Francisco, CA
Joined January 2017
- Tweets 138
- Following 517
- Followers 549
- Likes 3,431
Pinned Tweet
I'm excited to release what I've been cooking up the past few months at @arcinstitute
BINSEQ is a family of binary file formats for sequencing data built with paired records and parallel processing in mind with big performance gains (2x-40x) over gzip-fastq with similar storage
BINSEQ: A Family of High-Performance Binary Formats for Nucleotide Sequences biorxiv.org/content/10.1101/… #biorxiv_bioinfo
And stay on the look out the next couple weeks (hopefully) for the release of an even bigger project built with binseq!
And if you're interested in building with binseq here is the place to start!
github.com/arcinstitute/bins…
I've also added some nice functionality to bqtools including a very useful colored grep!
github.com/arcinstitute/bqto…
I've updated the BINSEQ manuscript to stay up to date with changes since I originally put it out at the beginning of the year
Some notable changes:
1. Support for ambiguous bases with 4bit encoding
2. Support for sequence headers
3. Improved API
biorxiv.org/content/10.1101/…
Noam Teyssier retweeted
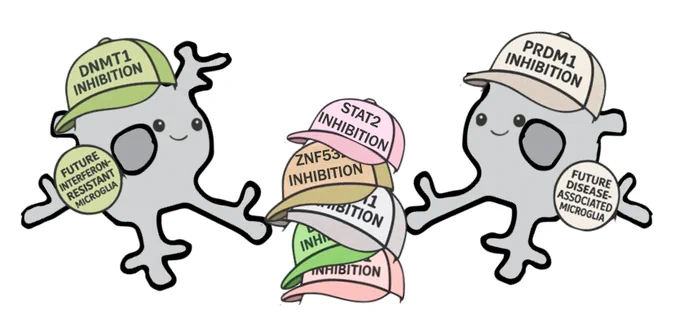
I’m super excited to announce our preprint on Transcriptional regulation of disease-relevant microglial activation programs. To determine regulators of microglia activation states, we performed six transcription factor-wide CRISPRi screens. biorxiv.org/content/10.1101/…
Noam Teyssier retweeted
🌎👩🔬 For 15+ years biology has accumulated petabytes (million gigabytes) of🧬DNA sequencing data🧬 from the far reaches of our planet.🦠🍄🌵
Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024.07.30.6…
Noam Teyssier retweeted
writing CUDA kernels is fun.
getting them to actually ship is pain.
we built kernel-builder so you can skip the pain → huggingface.co/blog/kernel-b…
Noam Teyssier retweeted
Computing x engineering biology is a playground for systems. Come join us for round three of the SF systems reading group with a focus on biotech.
- Noam Teyssier (Arc Institute): BINSEQ: A Family of High Performance Binary Formats for Nucleotide Sequences
- Aidan Abdullali (LatchBio): A Distributed Filesystem Built on Postgres and S3
- Abhinav Adduri (Arc Institute): Scaling Deep Learning to 1B+ Single Cells
Presentations on design decisions and paper highlights. We'll read snippets of source code, learn from each other and vibe. 5:30ish on 8/20
I use this all the time! Great for quick inspections without loading up a jupyter notebook
I actually submitted a PR to uv a while back so the `--with` flag accepts a CSV list so you can run a simple oneliner
`uvx --with numpy,polars,seaborn ipython`
Noam Teyssier retweeted
Register today for the Virtual Cell Challenge and use AI to solve one of biology’s most complex problems.
Announced in @CellCellPress, the competition is hosted by Arc Institute and sponsored by @nvidia, @10xGenomics, and @UltimaGenomics.
Noam Teyssier retweeted
Cells are dynamic, messy and context dependent. Scaling models across diverse states needs flexibility to capture heterogeneity
Introducing State, a transformer that predicts perturbation effects by training over sets of cells
Team effort led by the unstoppable @abhinadduri
Noam Teyssier retweeted
Today @arcinstitute releases State, our first perturbation prediction AI model and an important step towards our goal of a virtual cell
State is designed to learn how to shift cells between states (e.g. “diseased” to “healthy”) using drugs, cytokines, or genetic perturbations
Noam Teyssier retweeted
Excited to share the Kernel Hub, optimized CUDA kernels, plug-and-play from the Hugging Face Hub.
No boilerplate, just speed.
huggingface.co/blog/hello-hf…
Noam Teyssier retweeted
Ten years after Rust 1.0, we need to stop thinking of it as just "systems programming." That label carries historic baggage and scares away teams who could benefit from Rust.
corrode.dev/blog/foundationa…
#rustlang #rust
Noam Teyssier retweeted
Slides from my talk (with Kamil Jaron) on an history of k-mers in bioinformatics: rayan.chikhi.name/pdf/2025-k…
Noam Teyssier retweeted
Genomes encode biological complexity, which is determined by combinations of DNA mutations across millions of bases
In new @arcinstitute work, we report the discovery and engineering of the first programmable DNA recombinases capable of megabase-scale human genome rearrangement
What if we could universally recombine, insert, delete, or invert any two pieces of DNA?
In back-to-back @Nature papers, we report the discovery of bridge RNAs and 3 atomic structures of the first natural RNA-guided recombinase - a new mechanism for programmable genome design
Noam Teyssier retweeted
CERN Scientists today:
Scientists at CERN's Large Hadron Collider have successfully transformed lead into gold atoms, achieving an ancient alchemist dream through modern physics. abcnews.link/zqp39oz
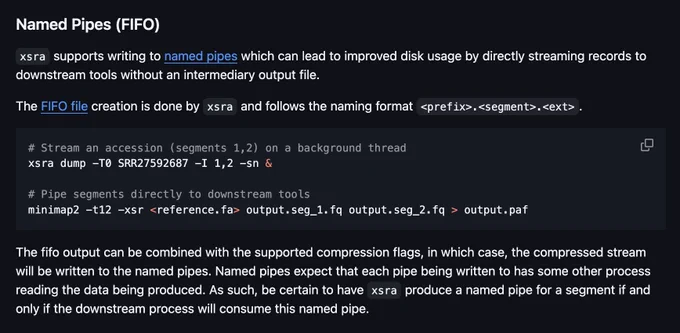
Thanks to Noam's clean code & Rust's readability, I was able to help Noam integrate this in short order! This is a feature I've long wanted from fastq-dump/fasterq-dump, it took xsra to get it! Stream multiple segments from xsra directly to your downstream preprocessing tools!
Just merged in a nice PR to xsra this morning to increase streaming support with named pipes (FIFO).
You can stream your R1/R2 directly to other tools and skip the intermediate write step.
It works with both on- and off-disk accessions
Give it a shot!
github.com/arcInstitute/xsra
Just merged in a nice PR to xsra this morning to increase streaming support with named pipes (FIFO).
You can stream your R1/R2 directly to other tools and skip the intermediate write step.
It works with both on- and off-disk accessions
Give it a shot!
github.com/arcInstitute/xsra