MD/PhD Computational Genomics, Johnson & Johnson
Joined June 2009
- Tweets 367
- Following 1,230
- Followers 208
- Likes 226
Mehdi Pirooznia retweeted
Interested in LLMs for genomic research but don't know where to start? looking for a review/survey to get started in this field? 👇👇😀
I am very excited to share that our review paper titled "To Transformers and Beyond: Large Language Models for the Genome" is now available as a preprint (arxiv.org/abs/2311.07621)! Our review unveils a revolution in genomics analysis with Genome LLMs. 🧬
🔍 What's Inside:
✅ The power & challenges of transformers in genomics.
✅ Cutting-edge models like HeynaDNA and scGPT & their impact.
✅ Deep dives into Enformer, DNABERT, and other Genome LLMs.
🌍 Why it Matters:
1. GPT-4's influence reshapes AI in genomics.
Unmatched insights into transformers' role in genomics.
2. Critical analysis of new models, addressing interpretability, privacy, & computational needs.
3. Essential for computational biologists & computer scientists to navigate the future of genomic data analysis.
This is a work led by the amazing PhD student, Mica Consens, in the lab! Also, a huge collaborative work with lots of field leaders @fabian_theis @genophoria @MKarimzade @michaelwainberg and Alan Moses!
@UofT @VectorInst @UofTCompSci @UofT_LMP @UHN @pmcc_ai @UHNAIHUB
Mehdi Pirooznia retweeted
Did @10xGenomics just make newer versions of Cell Ranger open source? github.com/10XGenomics/cellr… (cc @sjackman, @luizirber). This is a huge win for open, transparent, and reproducible science if it’s the case!
Mehdi Pirooznia retweeted
Computational immunogenomic approaches to predict response to cancer immunotherapies nature.com/articles/s41571-0…
Mehdi Pirooznia retweeted
We are happy to announce release of FinnGen-UKBB meta-analysis results for 679 phenotypes at:
public-metaresults-fg-ukbb.f…
- enjoy browsing and downloading!
#genomics #OpenScience
Mehdi Pirooznia retweeted
pyCirclize: a Python implementation of circlize
github.com/moshi4/pyCirclize
Mehdi Pirooznia retweeted
veloVI enhances RNA velocity analysis with uncertainty quantification and extensibility by deep generative modeling of gene-specific transcriptional dynamics. @adamgayoso @PhilippWeiler7 @fabian_theis @YosefLab
OA paper: nature.com/articles/s41592-0…
Mehdi Pirooznia retweeted
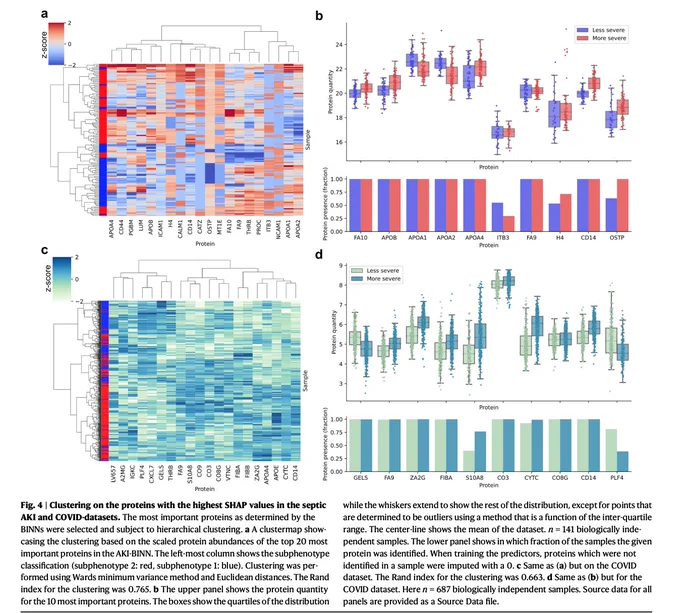
Interpreting biologically informed neural networks for enhanced proteomic biomarker discovery and pathway analysis | Nature Communications
nature.com/articles/s41467-0…
#Bioinformatics
Mehdi Pirooznia retweeted
Systematic transcriptional analysis of human cell lines for gene expression landscape and tumor representation🧬
nature.com/articles/s41467-0…
Mehdi Pirooznia retweeted
We're still learning about cancer immunotherapy
—When more mutations don't lead to a better response
nature.com/articles/s41588-0… @NatureGenet @PeterMKWestcott @LabJacks @isidrolauscher
—Outstanding review of targeting innate pathways
cell.com/immunity/fulltext/S… @ImmunityCP @jkagan1
Mehdi Pirooznia retweeted
Cellatlas: Universal preprocessing of single-cell genomics data biorxiv.org/content/10.1101/…
Mehdi Pirooznia retweeted
Automated Bioinformatics Analysis via AutoBA biorxiv.org/content/10.1101/…
Mehdi Pirooznia retweeted
Do you want to build a hierarchy of cell types using a single-cell reference Atlases? and the.use that to map a new query and detect novel cell-types/states, or harmonize cell-types across multiple datasets ? Then check treeArches academic.oup.com/nargab/arti……
Mehdi Pirooznia retweeted
SCROAM: Highly accurate estimation of cell type abundance in bulk tissues based on single-cell reference and domain adaptive matching biorxiv.org/content/10.1101/…
Mehdi Pirooznia retweeted
New #JITC article: Efficacy of PD-(L)1 blockade monotherapy compared with PD-(L)1 blockade plus chemotherapy in first-line PD-L1-positive advanced lung adenocarcinomas: a cohort study jitc.bmj.com/content/11/7/e0… @ArielleElkrief @alessi_joao @XinAnnWang @charlesrudin @DrMarkAwad @AdamJSchoenfeld
Mehdi Pirooznia retweeted
In case it is of help. With tidyHeatmap (uses ComplexHM as engine):
tbl |> heatmap(
sample, symbol, count,
scale = "row"
) |>
layer_point( pvalue<0.05 ) |>
layer_square(...) |>
layer_arrow_up(...)
github.com/stemangiola/tidyH…
Mehdi Pirooznia retweeted
Want to join a global consortium using #singlecell technologies to understand human diversity? Are #AI and #MachineLearning your #s? Come to Singapore! We're looking for leaders to drive v1 of the Human Diversity Atlas within the #HumanCellAtlas. tinyurl.com/5ccsfffz
Mehdi Pirooznia retweeted
EGFRm+ NSCLC treatment landscape
Key assets, trials and companies segmented by treatment line and mutation type
Some takeaways from this competitive space:
Mehdi Pirooznia retweeted
Clustering algorithms report clusters even when none exist. In single-cell RNA-Seq pipelines, novel cell types are often identified by clustering algorithms. Expanding on Kimes et al.'s work, we introduce significance analysis for single-cell RNA-Seq data: nature.com/articles/s41592-0…
Mehdi Pirooznia retweeted
This Fall I'll be teaching "quick data visualization" as part of a 1st year grad student seminar again for PBGG at UGA. The graphical abstract has 7 sections, but an 8th section on figure assembly has been added as well.
Contents available on GitHub: github.com/cxli233/Online_R_…